
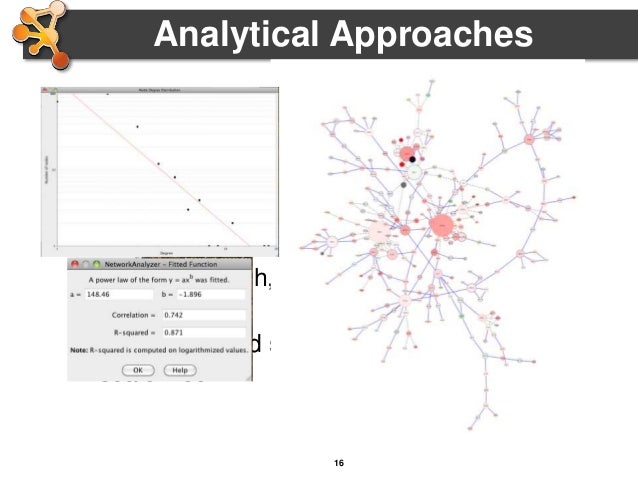
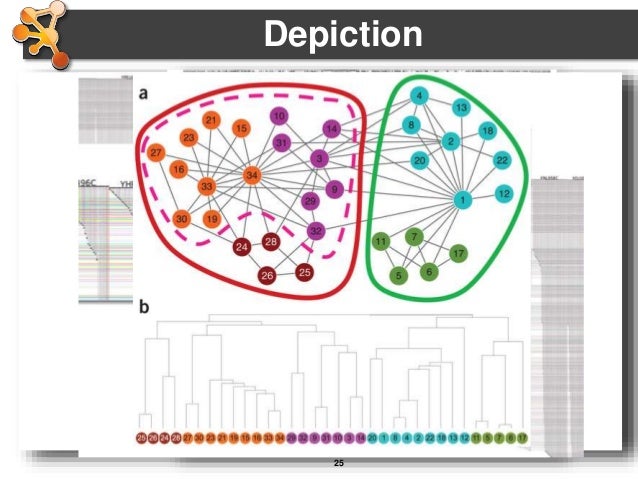
A targeted curation approach is necessary to transfer published data generated by primarily low-throughput techniques into interaction databases. Meaningfully representing this information remains far from trivial and different databases strive to provide users with detailed records capturing the experimental details behind each piece of interaction evidence. Network graphs generated from this data provide an understanding of the relationships between different proteins in the cell, and network analysis has become a widespread tool supporting -omics analysis. Molecular interaction databases are essential resources that enable access to a wealth of information on associations between proteins and other biomolecules. In this manuscript, we introduce these two algorithms and provide community access to the tool suite, describe examples of how these tools are useful to selectively present molecular interaction data and demonstrate a case where the algorithms were successfully used to identify a systematic error in an existing dataset. We have developed both a merging algorithm and a scoring system for molecular interactions based on the proteomics standard initiative-molecular interaction standards. Merging and scoring of data are commonly required operations after querying for the details of specific molecular interactions, to remove redundancy and assess the strength of accompanying experimental evidence. Experimental data is captured in multiple repositories, but there is no simple way to assess the evidence of an interaction occurring in a cellular environment.
Cytoscape 3 tutorial install#
I strongly recommend to use Docker to run your notebook server, but if you want to build on your machine, install the following to try these notebooks:Īgain, if you use the Docker container we provide, you don't have to install any of these.The evidence that two molecules interact in a living cell is often inferred from multiple different experiments. Docker is a powerful tool to make your workflow reproducible, but it is not the main forcus of this course.
Cytoscape 3 tutorial software#
About DockerĪll required software packages to run these lessons are packed in Docker container for your convenience. Building simple data analysis workflows requires only basic knowledge of Python. You don't have to worry too much about Python.


 0 kommentar(er)
0 kommentar(er)
